MLOmics: Cancer Multi-Omics Database for Machine Learning
Abstract
Framing the investigation of diverse cancers as a machine learning problem has recently shown significant potential in multi-omics analysis and cancer research. Empowering these successful machine learning models are the high-quality training datasets with sufficient data volume and adequate preprocessing. However, while there exist several public data portals, these databases are not off-the-shelf for existing machine learning models. In this paper, we introduce MLOmics, an open cancer multi-omics database aiming at serving better the development and evaluation of bioinformatics and machine learning models. MLOmics contains 8,314 patient samples covering all 32 cancer types with four omics types, stratified features, and extensive baselines. Complementary support for downstream analysis and bio-knowledge linking are also included to support interdisciplinary analysis.

Figure 1: Schematic workflow of creating the MLOmics. The process starts with collecting patient samples covering 32 cancer types from the TCGA project. All resources in diverse data types and sizes are uniformly integrated and processed to contain data of four omics types. Datasets for benchmark ML tasks were constructed based on the processed data. MLOmics also selected baselines, metrics, and resources to support downstream biological analysis. Overview of the MLOmics. MLOmics provides an interface for developing and evaluating machine learning models based on cancer multi-omics data. MLOmics provides datasets in three feature scales for 20 classification, clustering, and omics imputation learning tasks. MLOmics also provides statistical, ML, and DL baselines for each task, which are evaluated by fair metrics. Bio-knowledge database linking with MLOmics. MLOmics provides resources to link with other bio-knowledge databases, enabling the integration of outer resources for applications such as ML evaluation, gene-disease association exploration, network inference, and functional analysis.
Usage Note
# Select the original scale GS-BRCA dataset
# and train the DeepCC model for the classification task.
cd Scripts/Classification
./DeepCC.sh GS-BRCA Original
# Select the top scale ACC dataset
# and train the Subtype-GAN model for the clustering task.
cd Scripts/Clustering
./Subtype-GAN.sh ACC Top
# Select the top scale ACC dataset
# and train the GAIN model for the imputation task (0.3 missing rate).
cd Scripts/Imputation
#!/bin/bash
# Save the current directory
current_dir=$(pwd)
# Navigate to the specific folder
cd ../../../Classification_and_Clustering/Python/
Subtype-GAN
# Record the start time
echo "Script started at: $(date)" >> results/$1_$2.cc
# Set dummy number of clusters to 4
python SubtypeGAN.py -m SubtypeGAN -n 4 -t $1_$2
python SubtypeGAN.py -m cc -t $1_$2
# Record the end time
echo "Script ended at: $(date)" >> results/$1_$2.cc
# Navigate back to the original directory
cd "$current_dir"
# Differential gene expression analysis (P-value = 0.05) with volcano plot.
cd Scripts/Dwonstream_Analysis
./volcano.sh <clustering_log_path> [options]
--p_value_threshold 0.05
# Pathway enrichment analysis (P-value = 0.05)
# with KEGG Pathway visualization.
cd Scripts/Dwonstream_Analysis
./pwanalysis.sh <clustering_log_path> [options]
--p_value_cutoff 0.05
# <clustering_log_path>: Location of resultant sample label.
# [options]: Optional parameters such as P-value (e.g.,
# p_value_threshold 0.05)
Experimental results and downstream analyses
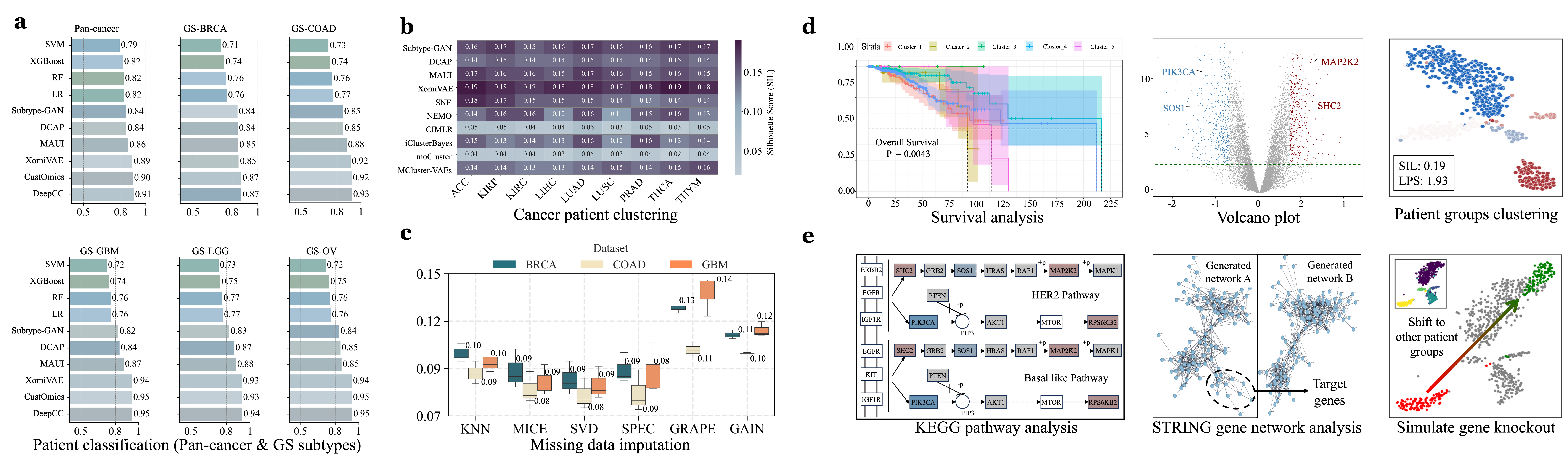
(a) Precision bar plots for each baseline method across all datasets. (b) SIL heatmaps for each baseline method across all datasets. (c) Box plots for each baseline method across three imputation datasets.(d–e) Schematic illustrations of downstream analysis results based on the clustering outcomes of a ML model applied to specific cancer patient clustering datasets.
Reference
Please kindly cite our paper if you use our datasets or results:@article{2025mlomics,
title = {MLOmics: Cancer Multi-Omics Database for Machine Learning},
author = {Yang, Ziwei and Kotoge, Rikuto and Piao, Xihao and Chen, Zheng and Zhu, Lingwei and Gao, Peng and Matsubara, Yasuko and Sakurai, Yasushi and Sun, Jimeng},
journal = {Scientific Data},
pages = {1--9},
year = {2025},
publisher = {Nature Publishing Group}
}